download.file(
url = "https://github.com/jhelvy/p4a/raw/main/data/wildlife_impacts.csv",
destfile = file.path('data', 'wildlife_impacts.csv')
)12 Data Wrangling
Learning Objectives
- Describe the purpose of the
dplyrpackage.- Select certain columns in a data frame with the
selectfunction.- Select certain rows in a data frame according to filtering conditions with the
filterfunction.- Link the output of one
dplyrfunction to the input of another function with the “pipe” operator%>%.- Add new columns to a data frame that are functions of existing columns with the
mutatefunction.- Sort data frames using the
arrange()function.- Use the split-apply-combine concept for data analysis.
- Use
summarize,group_by, andcountto split a data frame into groups of observations, apply summary statistics for each group, and then combine the results.- Export a data frame to a .csv file.
Suggested Readings
- Chapter 5 of “R for Data Science”, by Garrett Grolemund and Hadley Wickham
dplyrcheatsheet
12.1 R Setup
Before we get started, let’s set up our analysis environment:
- Open up your “data-analysis-tutorial” R Project that you created in the last lesson - if you didn’t do this, go back and do it now.
- Create a new
.Rfile (File > New File > R Script), and save it as “data_wrangling.R” inside your “data-analysis-tutorial” R Project folder. - Use the
download.file()function to download thewildlife_impacts.csvdataset, and save it in thedatafolder in your R Project:
For this lesson, we are going to use the FAA Wildlife Strike Database, which contains records of reported wildlife strikes with aircraft since 1990. Since aircraft-wildlife impacts are voluntarily reported, the database only contains information from airlines, airports, pilots, and other sources and does not represent all strikes. Each row in the dataset holds information for a single strike event with the following columns:
| Variable | Class | Description |
|---|---|---|
| incident_date | date | Date of incident |
| state | character | State |
| airport_id | character | ICAO Airport ID |
| airport | character | Airport Name |
| operator | character | Operator/Airline |
| atype | character | Airline type |
| type_eng | character | Engine type |
| species_id | character | Species ID |
| species | character | Species |
| damage | character | Damage: N None M Minor, M Uncertain, S Substantial, D Destroyed |
| num_engs | character | Number of engines |
| incident_month | double | Incident month |
| incident_year | double | Incident year |
| time_of_day | character | Incident Time of day |
| time | double | Incident time |
| height | double | Plane height at impact (feet) |
| speed | double | Plane speed at impact (knots) |
| phase_of_flt | character | Phase of flight at impact |
| sky | character | Sky condition |
| precip | character | Precipitation |
| cost_repairs_infl_adj | double | Cost of repairs adjusted for inflation |
Let’s load our libraries and read in the data:
library(readr)
library(dplyr)
df <- read_csv(file.path('data', 'wildlife_impacts.csv'))Just like in the last lesson, a good starting point when working with a new dataset is to view some quick summaries. Here’s another summary function (glimpse()) that is similar to str():
glimpse(df)#> Rows: 56,978
#> Columns: 21
#> $ incident_date <dttm> 2018-12-31, 2018-12-29, 2018-12-29, 2018-12-…
#> $ state <chr> "FL", "IN", "N/A", "N/A", "N/A", "FL", "FL", …
#> $ airport_id <chr> "KMIA", "KIND", "ZZZZ", "ZZZZ", "ZZZZ", "KMIA…
#> $ airport <chr> "MIAMI INTL", "INDIANAPOLIS INTL ARPT", "UNKN…
#> $ operator <chr> "AMERICAN AIRLINES", "AMERICAN AIRLINES", "AM…
#> $ atype <chr> "B-737-800", "B-737-800", "UNKNOWN", "B-737-9…
#> $ type_eng <chr> "D", "D", NA, "D", "D", "D", "D", "D", "D", "…
#> $ species_id <chr> "UNKBL", "R", "R2004", "N5205", "J2139", "UNK…
#> $ species <chr> "Unknown bird - large", "Owls", "Short-eared …
#> $ damage <chr> "M?", "N", NA, "M?", "M?", "N", "N", "N", "N"…
#> $ num_engs <dbl> 2, 2, NA, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,…
#> $ incident_month <dbl> 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 1…
#> $ incident_year <dbl> 2018, 2018, 2018, 2018, 2018, 2018, 2018, 201…
#> $ time_of_day <chr> "Day", "Night", NA, NA, NA, "Day", "Night", N…
#> $ time <dbl> 1207, 2355, NA, NA, NA, 955, 948, NA, NA, 132…
#> $ height <dbl> 700, 0, NA, NA, NA, NA, 600, NA, NA, 0, NA, 0…
#> $ speed <dbl> 200, NA, NA, NA, NA, NA, 145, NA, NA, 130, NA…
#> $ phase_of_flt <chr> "Climb", "Landing Roll", NA, NA, NA, "Approac…
#> $ sky <chr> "Some Cloud", NA, NA, NA, NA, NA, "Some Cloud…
#> $ precip <chr> "None", NA, NA, NA, NA, NA, "None", NA, NA, "…
#> $ cost_repairs_infl_adj <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…Wow, there have been 56,978 reported wildlife strikes over the 29 period from 1990 to 2019! On a daily average that comes out to:
nrow(df) / (2019 - 1990) / 365#> [1] 5.3829…over 5 strikes per day!
12.2 Data wrangling with dplyr

“Data Wrangling” refers to the art of getting your data into R in a useful form for visualization and modeling. Wrangling is the first step in the general data science process:
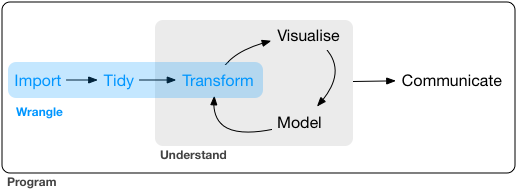
12.2.1 What is dplyr
As we saw in the last section, we can use brackets ([]) to access elements of a data frame. While this is handy, it can be cumbersome and difficult to read, especially for complicated operations.
Enter dplyr
The dplyr package was designed to make tabular data wrangling easier to perform and read. It pairs nicely with other libraries, such as ggplot2 for visualizing data (which we’ll cover next week). Together, dplyr, ggplot2, and a handful of other packages make up what is known as the “Tidyverse” - an opinionated collection of R packages designed for data science. You can load all of the tidyverse packages at once using the library(tidyverse) command, but for now we’re just going to install and use each package one at a time - starting with dplyr:
install.packages("dplyr")In this lesson, we are going to learn some of the most common dplyr functions:
select(): subset columnsfilter(): subset rows on conditionsmutate(): create new columns by using information from other columnsarrange(): sort resultsgroup_by(): group data to perform grouped operationssummarize(): create summary statistics (usually on grouped data)count(): count discrete rows
12.2.2 Select columns with select()
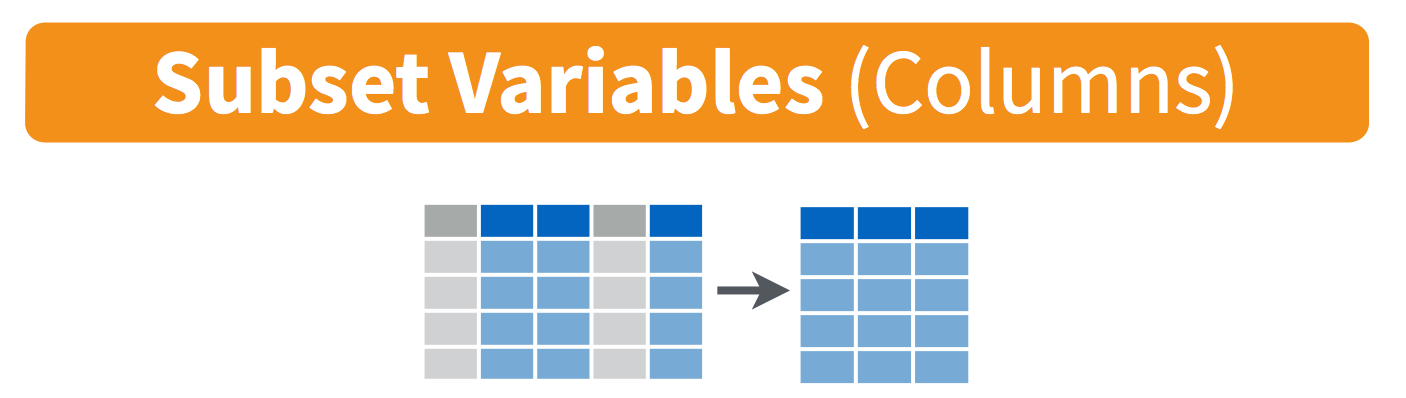
To select specific columns, use select(). The first argument to this function is the data frame (df), and the subsequent arguments are the columns to keep:
# Select only a few columns
select(df, state, damage, time_of_day)#> # A tibble: 56,978 × 3
#> state damage time_of_day
#> <chr> <chr> <chr>
#> 1 FL M? Day
#> 2 IN N Night
#> 3 N/A <NA> <NA>
#> 4 N/A M? <NA>
#> 5 N/A M? <NA>
#> 6 FL N Day
#> # ℹ 56,972 more rowsTo select all columns except certain ones, put a - sign in front of the variable to exclude it:
select(df, -state, -damage, -time_of_day)#> # A tibble: 56,978 × 18
#> incident_date airport_id airport operator atype type_eng species_id
#> <dttm> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 2018-12-31 00:00:00 KMIA MIAMI IN… AMERICA… B-73… D UNKBL
#> 2 2018-12-29 00:00:00 KIND INDIANAP… AMERICA… B-73… D R
#> 3 2018-12-29 00:00:00 ZZZZ UNKNOWN AMERICA… UNKN… <NA> R2004
#> 4 2018-12-27 00:00:00 ZZZZ UNKNOWN AMERICA… B-73… D N5205
#> 5 2018-12-27 00:00:00 ZZZZ UNKNOWN AMERICA… B-73… D J2139
#> 6 2018-12-27 00:00:00 KMIA MIAMI IN… AMERICA… A-319 D UNKB
#> # ℹ 56,972 more rows
#> # ℹ 11 more variables: species <chr>, num_engs <dbl>, incident_month <dbl>,
#> # incident_year <dbl>, time <dbl>, height <dbl>, speed <dbl>,
#> # phase_of_flt <chr>, sky <chr>, precip <chr>, cost_repairs_infl_adj <dbl>Some additional options to select columns based on a specific criteria include:
ends_with()= Select columns that end with a character stringcontains()= Select columns that contain a character stringmatches()= Select columns that match a regular expressionone_of()= Select column names that are from a group of names
12.2.3 Select rows with filter()

Filter the rows for wildlife impacts that occurred in DC:
filter(df, state == 'DC')#> # A tibble: 1,228 × 21
#> incident_date state airport_id airport operator atype type_eng
#> <dttm> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 2018-10-23 00:00:00 DC KDCA RONALD REAGAN… AMERICA… B-73… D
#> 2 2018-10-17 00:00:00 DC KDCA RONALD REAGAN… AMERICA… B-73… D
#> 3 2018-10-16 00:00:00 DC KDCA RONALD REAGAN… AMERICA… B-73… D
#> 4 2018-10-12 00:00:00 DC KDCA RONALD REAGAN… AMERICA… B-73… D
#> 5 2018-09-04 00:00:00 DC KDCA RONALD REAGAN… AMERICA… A-319 D
#> 6 2018-09-01 00:00:00 DC KDCA RONALD REAGAN… AMERICA… A-321 D
#> # ℹ 1,222 more rows
#> # ℹ 14 more variables: species_id <chr>, species <chr>, damage <chr>,
#> # num_engs <dbl>, incident_month <dbl>, incident_year <dbl>,
#> # time_of_day <chr>, time <dbl>, height <dbl>, speed <dbl>,
#> # phase_of_flt <chr>, sky <chr>, precip <chr>, cost_repairs_infl_adj <dbl>Filter the rows for wildlife impacts that cost more than $1 million in damages:
filter(df, cost_repairs_infl_adj > 10^6)#> # A tibble: 41 × 21
#> incident_date state airport_id airport operator atype type_eng
#> <dttm> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 2015-05-26 00:00:00 N/A SVMI SIMON BOLIVAR… AMERICA… B-73… D
#> 2 2015-04-25 00:00:00 FL KJAX JACKSONVILLE … AMERICA… A-319 D
#> 3 2015-04-02 00:00:00 MA KBOS GENERAL EDWAR… AMERICA… B-73… D
#> 4 2001-04-02 00:00:00 N/A LFPG CHARLES DE GA… AMERICA… B-76… D
#> 5 2015-10-08 00:00:00 WA KSEA SEATTLE-TACOM… DELTA A… A-330 D
#> 6 2015-09-23 00:00:00 UT KSLC SALT LAKE CIT… DELTA A… B-73… D
#> # ℹ 35 more rows
#> # ℹ 14 more variables: species_id <chr>, species <chr>, damage <chr>,
#> # num_engs <dbl>, incident_month <dbl>, incident_year <dbl>,
#> # time_of_day <chr>, time <dbl>, height <dbl>, speed <dbl>,
#> # phase_of_flt <chr>, sky <chr>, precip <chr>, cost_repairs_infl_adj <dbl>12.2.4 Sequence operations with pipes (%>%)

What if you want to select and filter at the same time? Well, one way to do this is to use intermediate steps. To do this, you first create a temporary data frame and then use that as input to the next function, like this:
dc_impacts <- filter(df, state == 'DC')
dc_impacts_airlineTime <- select(dc_impacts, operator, time, time_of_day)
head(dc_impacts_airlineTime)#> # A tibble: 6 × 3
#> operator time time_of_day
#> <chr> <dbl> <chr>
#> 1 AMERICAN AIRLINES 2130 Night
#> 2 AMERICAN AIRLINES 2043 Night
#> 3 AMERICAN AIRLINES 730 Dawn
#> 4 AMERICAN AIRLINES 2245 Night
#> 5 AMERICAN AIRLINES 2150 Night
#> 6 AMERICAN AIRLINES 2022 NightThis works, but it can also clutter up your workspace with lots of objects with different names.
Another approach is to use pipes, which is a more recent addition to R. Pipes let you take the output of one function and send it directly to the next, which is useful when you need to do many things to the same dataset.
The pipe operator is %>% and comes from the magrittr package, which is installed automatically with dplyr. If you use RStudio, you can type the pipe with Ctrl + Shift + M if you have a PC or Cmd + Shift + M if you have a Mac. Here’s the same thing as the previous example but with pipes:
df %>%
filter(state == 'DC') %>%
select(operator, time, time_of_day) %>%
head()#> # A tibble: 6 × 3
#> operator time time_of_day
#> <chr> <dbl> <chr>
#> 1 AMERICAN AIRLINES 2130 Night
#> 2 AMERICAN AIRLINES 2043 Night
#> 3 AMERICAN AIRLINES 730 Dawn
#> 4 AMERICAN AIRLINES 2245 Night
#> 5 AMERICAN AIRLINES 2150 Night
#> 6 AMERICAN AIRLINES 2022 NightIn the above code, we use the pipe to send the df data frame first through filter() to keep only rows from DC, and then through select() to keep only the columns operator, time, and time_of_day.
Since %>% takes the object on its left and passes it as the first argument to the function on its right, we don’t need to explicitly include the data frame as an argument to the filter() and select() functions.
Consider reading the %>% operator as the words “…and then…”. For instance, in the above example I would read the code as “First, filter to only data from DC, and then select the columns operator, time, and time_of_day, and then show the first 6 rows.”
Here’s another analogy:
Without Pipes:
leave_house(get_dressed(get_out_of_bed(wake_up(me))))With Pipes:
me %>%
wake_up %>%
get_out_of_bed %>%
get_dressed %>%
leave_houseIn the above example, adding pipes makes the flow of operations easier to read from left to right, with the %>% operator reading as “…and then…”
If you want to create a new object with the output of a “pipeline”, you just put the object name at the start of the first pipe:
dc_impacts <- df %>%
filter(state == 'DC') %>%
select(operator, time, time_of_day)
head(dc_impacts)#> # A tibble: 6 × 3
#> operator time time_of_day
#> <chr> <dbl> <chr>
#> 1 AMERICAN AIRLINES 2130 Night
#> 2 AMERICAN AIRLINES 2043 Night
#> 3 AMERICAN AIRLINES 730 Dawn
#> 4 AMERICAN AIRLINES 2245 Night
#> 5 AMERICAN AIRLINES 2150 Night
#> 6 AMERICAN AIRLINES 2022 Night12.2.5 Sort rows with arrange()
Use the arrange() function to sort a data frame by a column. For example, if you wanted to view the least expensive accidents, you could arrange the data frame by the variable cost_repairs_infl_adj:
# Arrange by least expensive accident
df %>%
arrange(cost_repairs_infl_adj)#> # A tibble: 56,978 × 21
#> incident_date state airport_id airport operator atype type_eng
#> <dttm> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 2013-09-05 00:00:00 MI KFNT BISHOP INTL A… SOUTHWE… B-73… D
#> 2 2011-04-17 00:00:00 TX KDFW DALLAS/FORT W… AMERICA… MD-80 D
#> 3 2018-07-10 00:00:00 NM KABQ ALBUQUERQUE I… SOUTHWE… B-73… D
#> 4 2017-10-31 00:00:00 PA KPIT PITTSBURGH IN… AMERICA… B-73… D
#> 5 2014-01-17 00:00:00 UT KSLC SALT LAKE CIT… SOUTHWE… B-73… D
#> 6 2006-04-28 00:00:00 TX KIAH GEORGE BUSH I… UNITED … B-73… D
#> # ℹ 56,972 more rows
#> # ℹ 14 more variables: species_id <chr>, species <chr>, damage <chr>,
#> # num_engs <dbl>, incident_month <dbl>, incident_year <dbl>,
#> # time_of_day <chr>, time <dbl>, height <dbl>, speed <dbl>,
#> # phase_of_flt <chr>, sky <chr>, precip <chr>, cost_repairs_infl_adj <dbl>To sort in descending order, add the desc() function inside the arrange() function. For example, here are the most expensive accidents:
# Arrange by most expensive accident
df %>%
arrange(desc(cost_repairs_infl_adj))#> # A tibble: 56,978 × 21
#> incident_date state airport_id airport operator atype type_eng
#> <dttm> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 2009-02-03 00:00:00 CO KDEN DENVER INTL A… UNITED … B-75… D
#> 2 2007-11-22 00:00:00 N/A LFMN NICE COTE D'A… DELTA A… B-76… D
#> 3 2011-09-26 00:00:00 CO KDEN DENVER INTL A… UNITED … B-75… D
#> 4 2017-07-11 00:00:00 CO KDEN DENVER INTL A… UNITED … B-73… D
#> 5 2008-10-25 00:00:00 UT KSLC SALT LAKE CIT… DELTA A… DC-9… D
#> 6 2011-07-30 00:00:00 CA KBUR BOB HOPE ARPT SOUTHWE… B-73… D
#> # ℹ 56,972 more rows
#> # ℹ 14 more variables: species_id <chr>, species <chr>, damage <chr>,
#> # num_engs <dbl>, incident_month <dbl>, incident_year <dbl>,
#> # time_of_day <chr>, time <dbl>, height <dbl>, speed <dbl>,
#> # phase_of_flt <chr>, sky <chr>, precip <chr>, cost_repairs_infl_adj <dbl>12.2.6 Create new variables with mutate()

You will often need to create new columns based on the values in existing columns. For this use mutate(). For example, let’s create a new variable converting the height variable from feet to miles:
df %>%
mutate(height_miles = height / 5280) %>%
select(height, height_miles)#> # A tibble: 56,978 × 2
#> height height_miles
#> <dbl> <dbl>
#> 1 700 0.133
#> 2 0 0
#> 3 NA NA
#> 4 NA NA
#> 5 NA NA
#> 6 NA NA
#> # ℹ 56,972 more rowsYou can also create a second new column based on the first new column within the same call of mutate():
df %>%
mutate(height_miles = height / 5280,
height_half_miles = height_miles / 2) %>%
select(height, height_miles, height_half_miles)#> # A tibble: 56,978 × 3
#> height height_miles height_half_miles
#> <dbl> <dbl> <dbl>
#> 1 700 0.133 0.0663
#> 2 0 0 0
#> 3 NA NA NA
#> 4 NA NA NA
#> 5 NA NA NA
#> 6 NA NA NA
#> # ℹ 56,972 more rowsYou’ll notice that the variables created have a lot of NAs - that’s because there are missing data. If you wanted to remove those, you could insert a filter() in the pipe chain:
df %>%
filter(!is.na(height)) %>%
mutate(height_miles = height / 5280) %>%
select(height, height_miles)#> # A tibble: 38,940 × 2
#> height height_miles
#> <dbl> <dbl>
#> 1 700 0.133
#> 2 0 0
#> 3 600 0.114
#> 4 0 0
#> 5 0 0
#> 6 0 0
#> # ℹ 38,934 more rowsis.na() is a function that determines whether something is an NA. The ! symbol negates the result, so we’re asking for every row where weight is not an NA.
12.3 Split-apply-combine
Many data analysis tasks can be approached using the split-apply-combine paradigm:
- Split the data into groups
- Apply some analysis to each group
- Combine the results.
dplyr makes this very easy through the use of the group_by() function.
12.3.1 The group_by() function
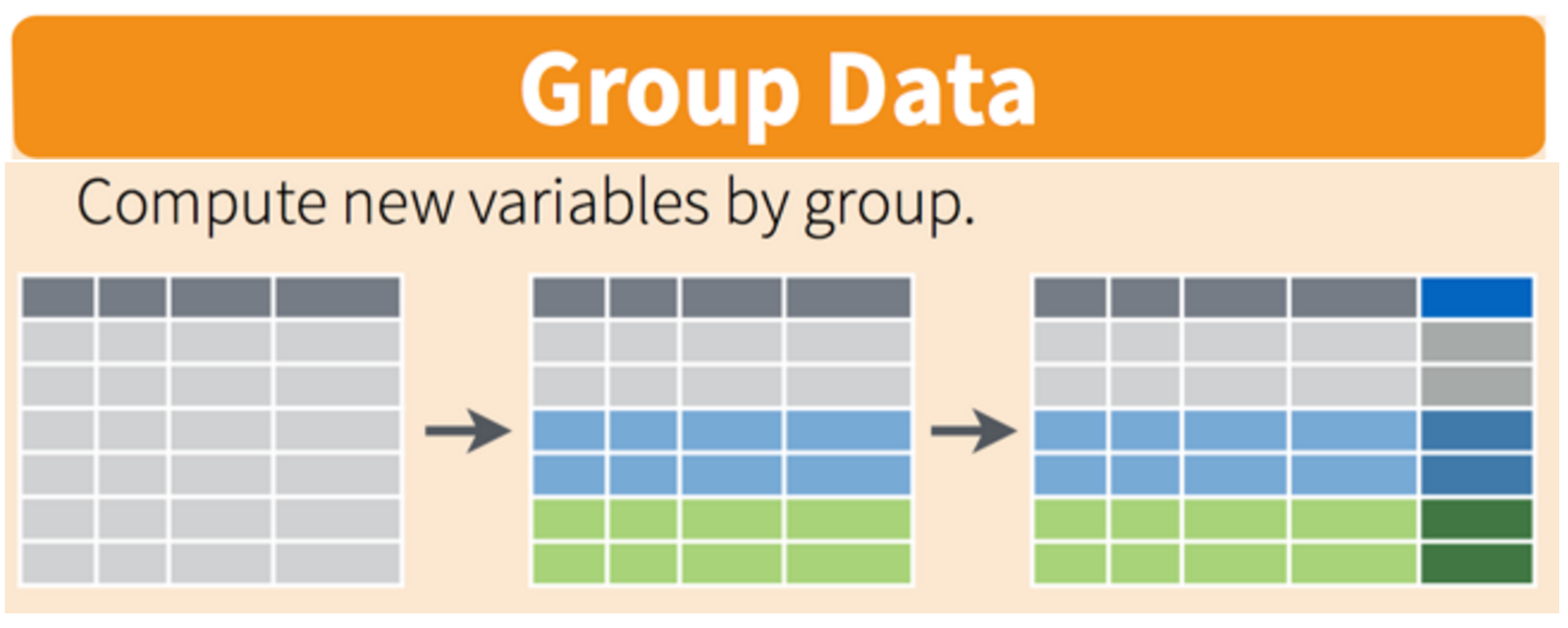
The group_by() function enables you to perform operations across groups within the data frame. It is typically used by inserting it in the “pipeline” before the desired group operation. For example, if we wanted to add a a new column that computed the mean height of reported wildlife impacts for each state, we could insert group_by(state) in the pipeline:
df %>%
filter(!is.na(height)) %>%
group_by(state) %>% # Here we're grouping by state
mutate(mean_height = mean(height)) %>%
select(state, mean_height)#> # A tibble: 38,940 × 2
#> # Groups: state [59]
#> state mean_height
#> <chr> <dbl>
#> 1 FL 892.
#> 2 IN 719.
#> 3 FL 892.
#> 4 FL 892.
#> 5 TX 1177.
#> 6 NY 937.
#> # ℹ 38,934 more rowsYou’ll see that the same value for mean_height is reported for the same states (e.g. the mean height in Florida is 892 ft).
12.3.2 The summarize() function
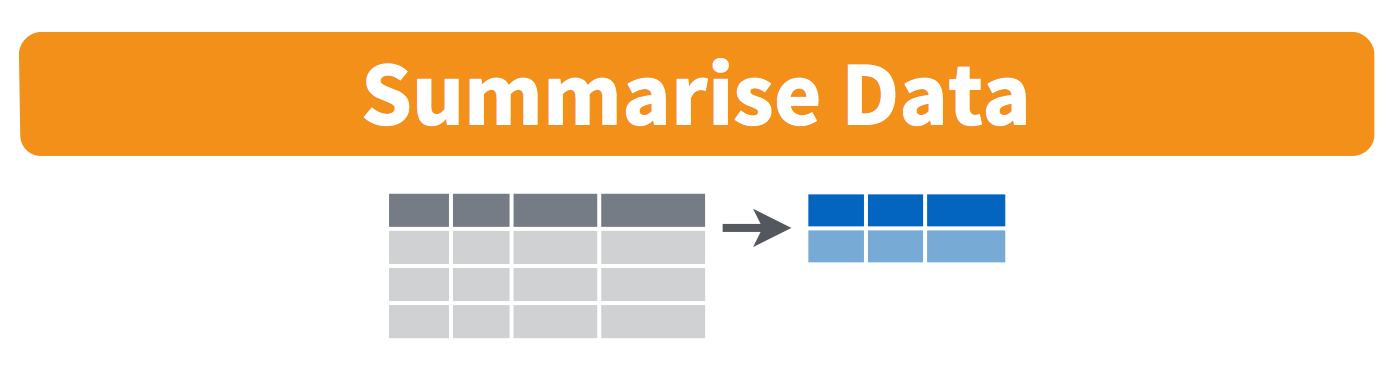
The group_by() function is often used together with summarize(), which collapses each group into a single-row summary of that group. For example, we collapse the result of the previous example by using summarise() instead of mutate():
df %>%
filter(!is.na(height)) %>%
group_by(state) %>%
summarise(mean_height = mean(height))#> # A tibble: 59 × 2
#> state mean_height
#> <chr> <dbl>
#> 1 AB 327.
#> 2 AK 334.
#> 3 AL 861.
#> 4 AR 968.
#> 5 AZ 2134.
#> 6 BC 497.
#> # ℹ 53 more rowsYou can also group by multiple columns - here let’s group by state and the airline:
df %>%
filter(!is.na(height)) %>%
group_by(state, operator) %>%
summarise(mean_height = mean(height))#> `summarise()` has grouped output by 'state'. You can override using the
#> `.groups` argument.#> # A tibble: 213 × 3
#> # Groups: state [59]
#> state operator mean_height
#> <chr> <chr> <dbl>
#> 1 AB AMERICAN AIRLINES 318.
#> 2 AB UNITED AIRLINES 350
#> 3 AK AMERICAN AIRLINES 0
#> 4 AK DELTA AIR LINES 414.
#> 5 AK UNITED AIRLINES 311.
#> 6 AL AMERICAN AIRLINES 1038.
#> # ℹ 207 more rowsNotice that in the above examples I’ve kept the early filter to drop NAs. This is important when performing summarizing functions like mean() or sum(). If NAs are present, the result will also be NA:
df %>%
group_by(state) %>%
summarise(mean_height = mean(height))#> # A tibble: 59 × 2
#> state mean_height
#> <chr> <dbl>
#> 1 AB NA
#> 2 AK 334.
#> 3 AL NA
#> 4 AR NA
#> 5 AZ NA
#> 6 BC NA
#> # ℹ 53 more rowsOnce the data are grouped, you can also summarize multiple variables at the same time (and not necessarily on the same variable). For instance, you could add two more columns computing the minimum and maximum height:
df %>%
filter(!is.na(height)) %>%
group_by(state, operator) %>%
summarise(mean_height = mean(height),
min_height = min(height),
max_height = max(height))#> `summarise()` has grouped output by 'state'. You can override using the
#> `.groups` argument.#> # A tibble: 213 × 5
#> # Groups: state [59]
#> state operator mean_height min_height max_height
#> <chr> <chr> <dbl> <dbl> <dbl>
#> 1 AB AMERICAN AIRLINES 318. 0 1300
#> 2 AB UNITED AIRLINES 350 0 1400
#> 3 AK AMERICAN AIRLINES 0 0 0
#> 4 AK DELTA AIR LINES 414. 0 1700
#> 5 AK UNITED AIRLINES 311. 0 1200
#> 6 AL AMERICAN AIRLINES 1038. 0 11300
#> # ℹ 207 more rows12.3.3 Counting
Often times you will want to know the number of observations found for each variable or combination of variables. One way to do this is to use the group_by() and summarise() functions in combination. For example, here is the number of observations for each aircraft engine type:
df %>%
group_by(type_eng) %>%
summarise(count = n())#> # A tibble: 5 × 2
#> type_eng count
#> <chr> <int>
#> 1 A 2
#> 2 C 34
#> 3 D 56705
#> 4 F 3
#> 5 <NA> 234Since this is such a common task, dplyr provides the count() function to do the same thing:
df %>%
count(type_eng)#> # A tibble: 5 × 2
#> type_eng n
#> <chr> <int>
#> 1 A 2
#> 2 C 34
#> 3 D 56705
#> 4 F 3
#> 5 <NA> 234For convenience, count() also provides the sort argument:
df %>%
count(type_eng, sort = TRUE)#> # A tibble: 5 × 2
#> type_eng n
#> <chr> <int>
#> 1 D 56705
#> 2 <NA> 234
#> 3 C 34
#> 4 F 3
#> 5 A 2You can also count the combination of variables by providing more than one column name to count():
df %>%
count(type_eng, num_engs, sort = TRUE)#> # A tibble: 10 × 3
#> type_eng num_engs n
#> <chr> <dbl> <int>
#> 1 D 2 53652
#> 2 D 3 2753
#> 3 D 4 299
#> 4 <NA> NA 232
#> 5 C 2 34
#> 6 F 2 3
#> # ℹ 4 more rowsHmm, looks like most reported wildlife impacts involve planes with 2 D-type engines.
12.4 Exporting data
Now that you have learned how to use dplyr to extract information from or summarize your raw data, you may want to export these new data sets. Similar to the read_csv() function used for reading CSV files into R, there is a write_csv() function that generates CSV files from data frames.
Important: Before using write_csv(), create a new folder called “data_output” and put it in your R Project folder. In general, you should never write generated datasets in the same directory as your raw data. The “data” folder should only contain the raw, unaltered data, and should be left alone to make sure we don’t delete or modify it.
Let’s save one of the summary data frames from the earlier examples where we computed the min, mean, and max heights of impacts by each state and airline:
heightSummary <- df %>%
filter(!is.na(height)) %>%
group_by(state, operator) %>%
summarise(mean_height = mean(height),
min_height = min(height),
max_height = max(height))#> `summarise()` has grouped output by 'state'. You can override using the
#> `.groups` argument.Save the the new heightSummary data frame as a CSV file in your “data_output” folder:
write_csv(heightSummary, path = file.path('data_output', 'heightSummary.csv')12.5 Tips
You will often need to create new variables based on a condition. To do this, you can use the if_else() function. Here’s the general syntax:
if_else(<condition>, <if TRUE>, <else>)The first argument is a condition. If the condition is TRUE, then the value given to the second argument will be used; if not, then the third argument value will be used.
Here’s an example of creating a variable to determine which months in the wildlife impacts data are in the summer:
df %>%
mutate(
summer_month = if_else(incident_month %in% c(6, 7, 8), TRUE, FALSE))Of course, in this particular case the if_else() function isn’t even needed because the condition returns TRUE and FALSE values. However, if you wanted to extend this example to determine all four seasons, you could use a series of nested if_else() functions:
df %>%
mutate(season = if_else(
incident_month %in% c(3, 4, 5), 'Spring', if_else(
incident_month %in% c(6, 7, 8), 'Summer', if_else(
incident_month %in% c(9, 10, 11), 'Fall', 'Winter')
)))Note: The Base R version of this function is ifelse(), but I recommend using the dplyr version, if_else(), as it is a stricter function.
Page sources
Some content on this page has been modified from other courses, including:
- Data Analysis and Visualization in R for Ecologists, by François Michonneau & Auriel Fournier. Zenodo: http://doi.org/10.5281/zenodo.3264888
- The amazing illustrations by Allison Horst